User Guide
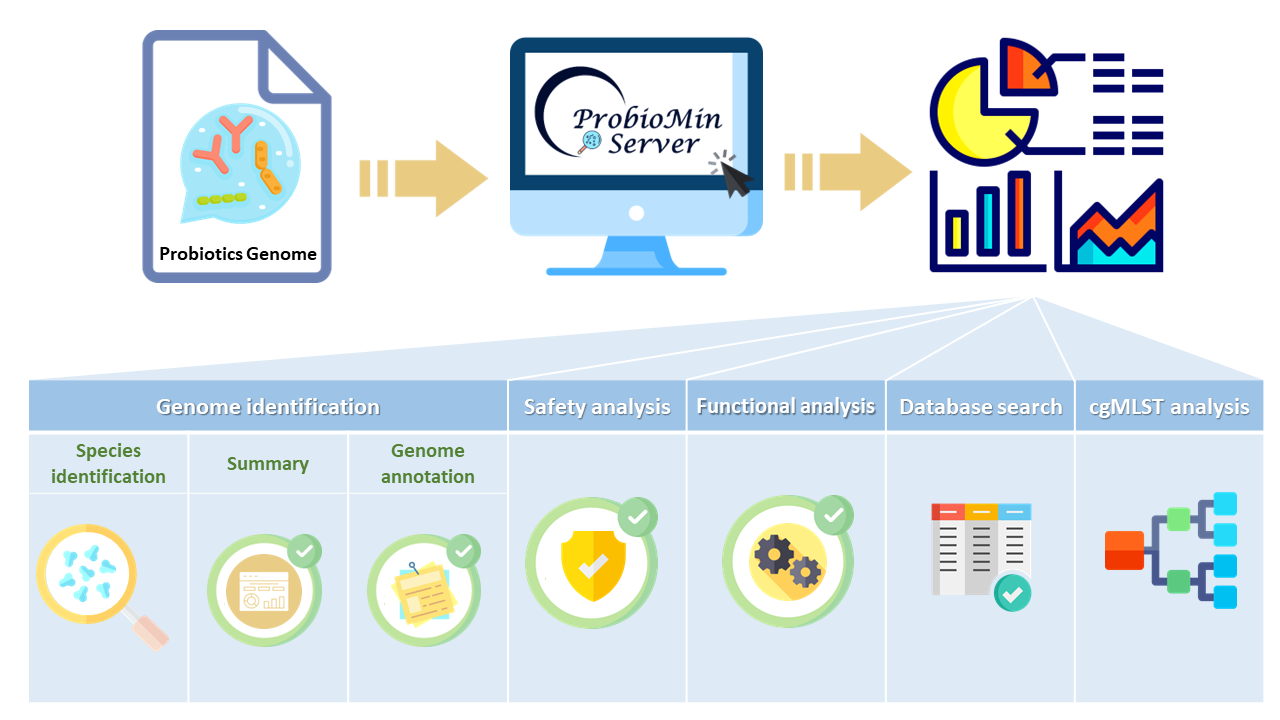
Graphical Abstract
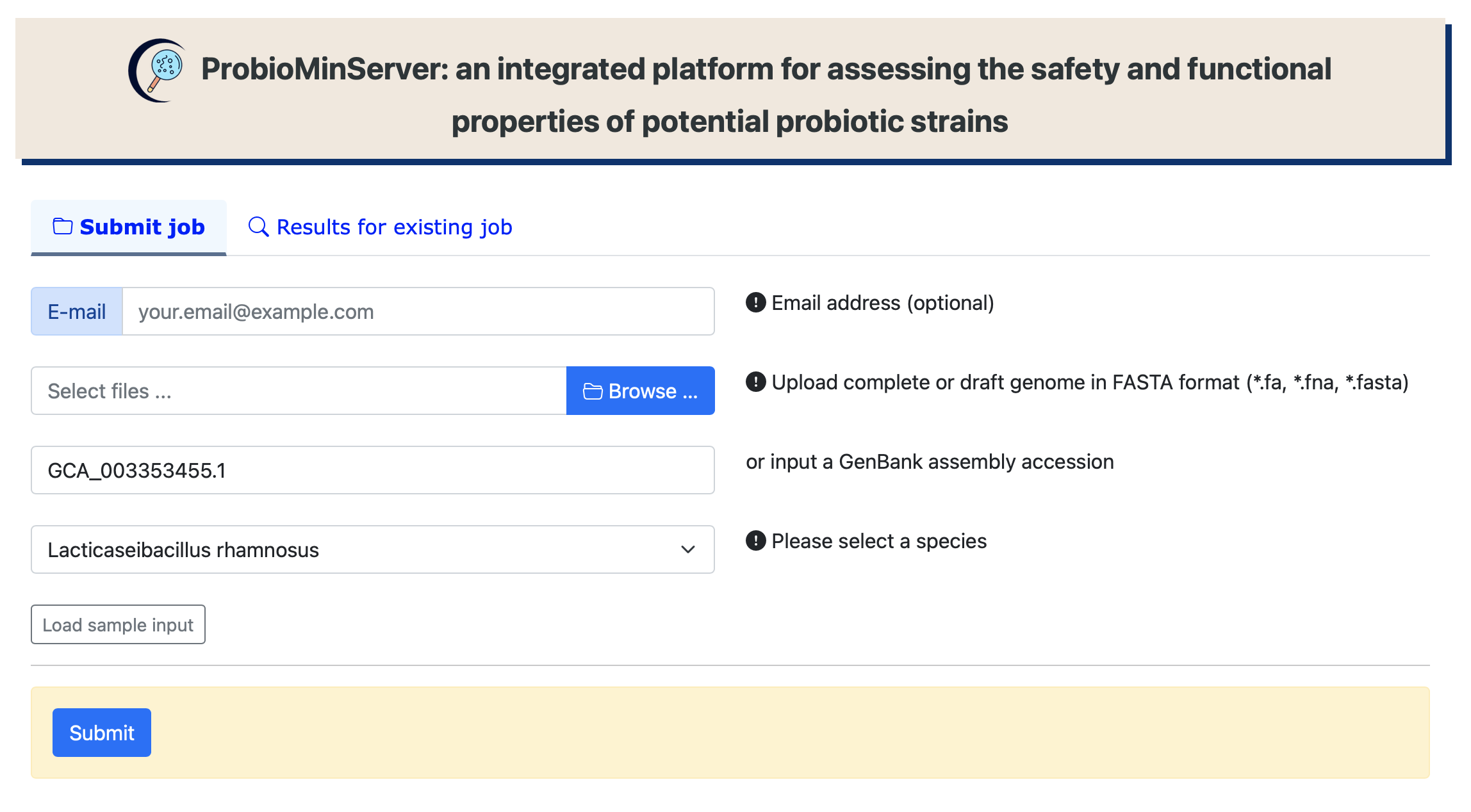
(I)ANALYSIS
Upload complete or draft genome file
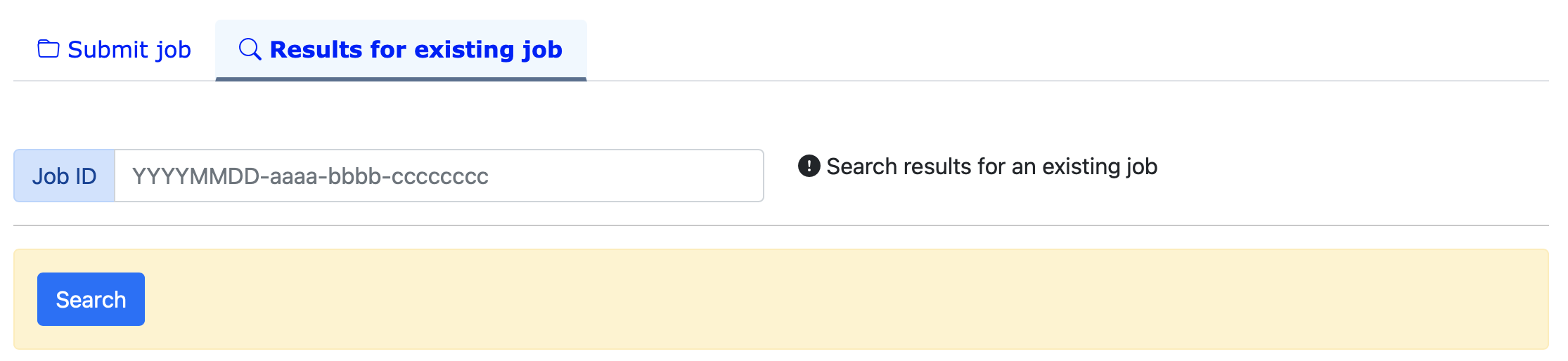
Search results for an existing job
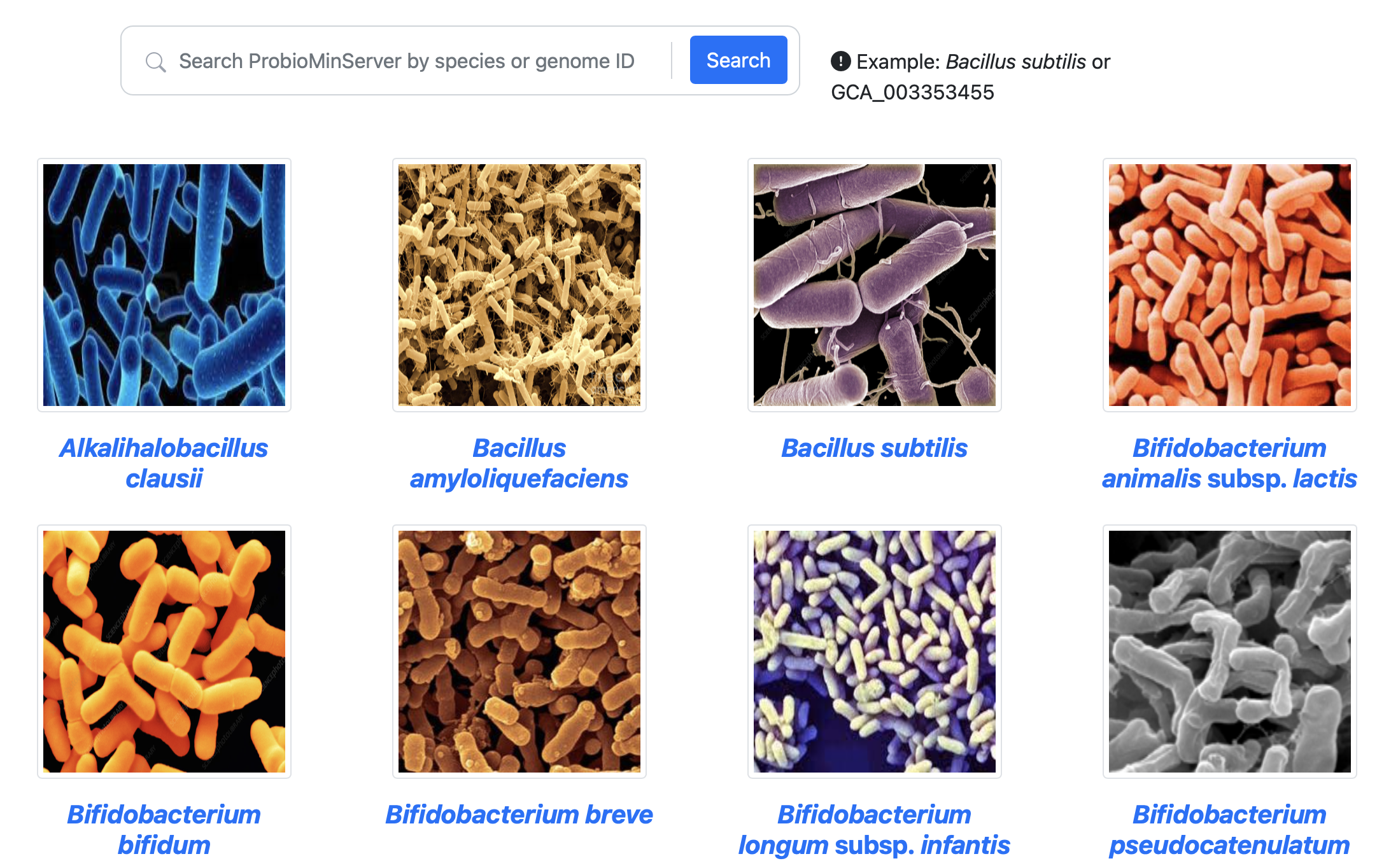
(II)BROWSE
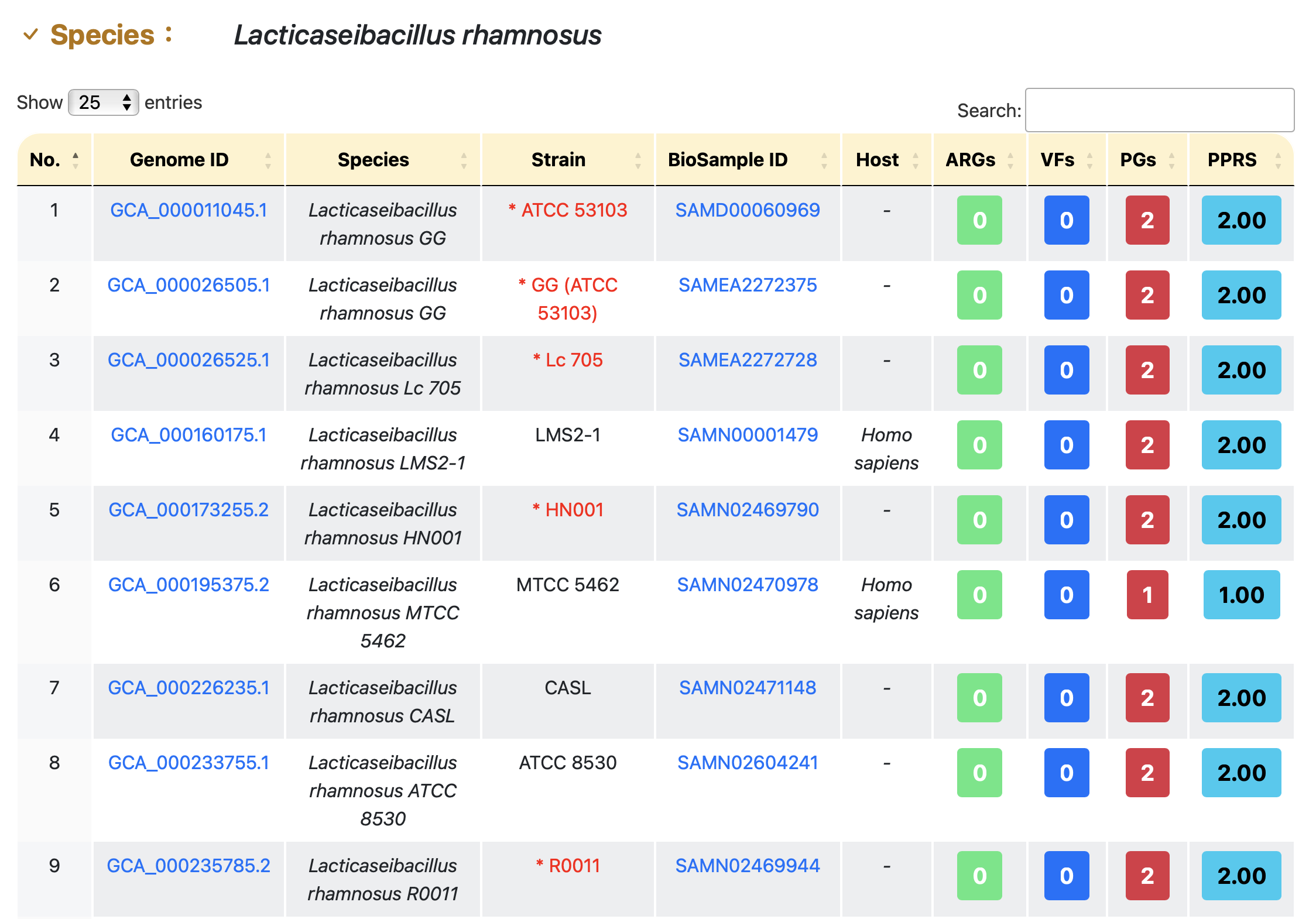
Search and browse database
Detail of a species
(III)RESULTS
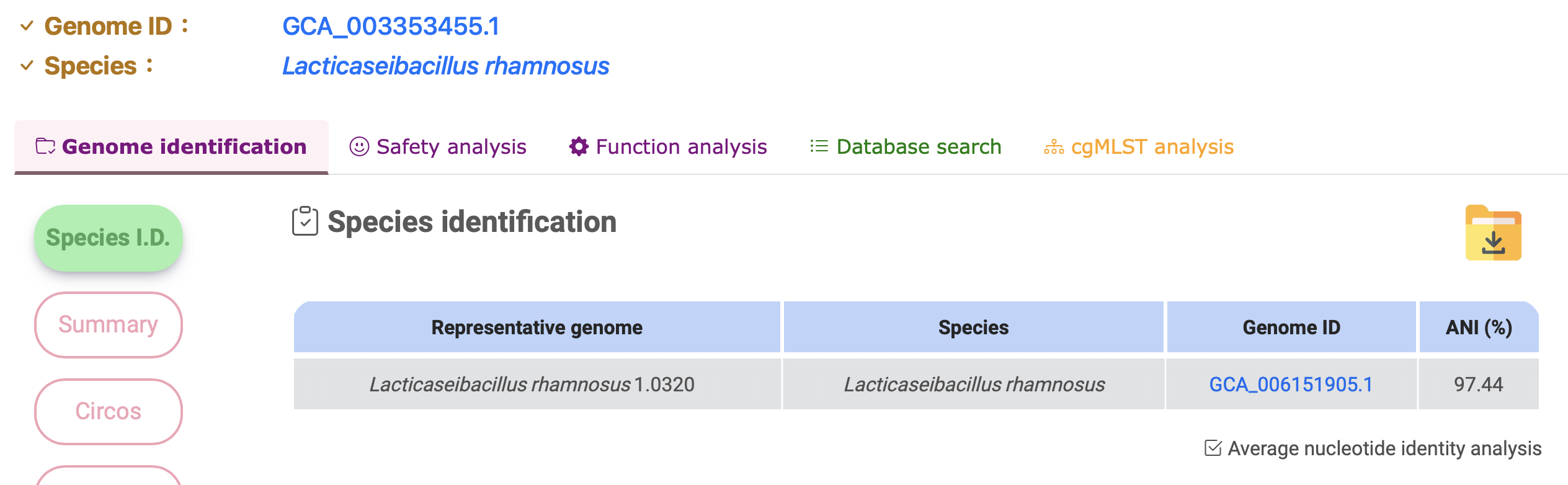
Species identification
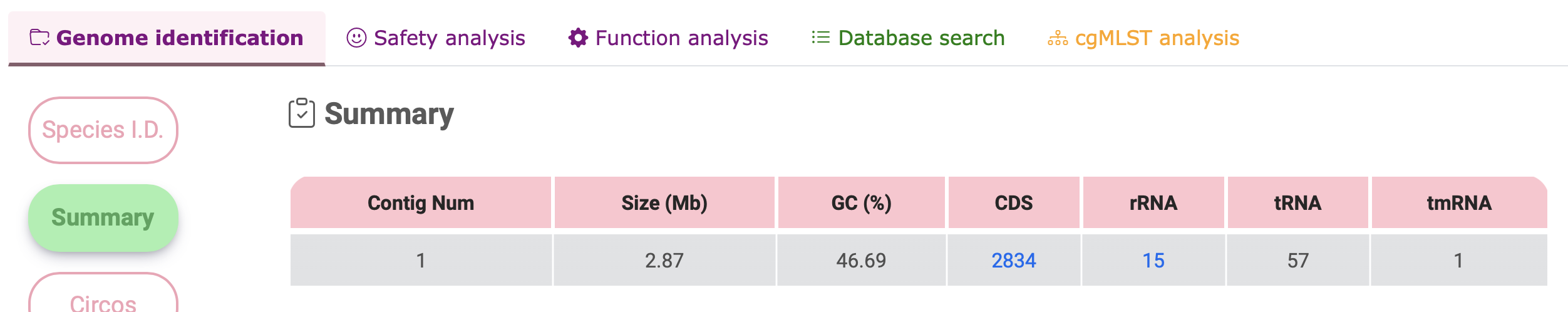
Summary
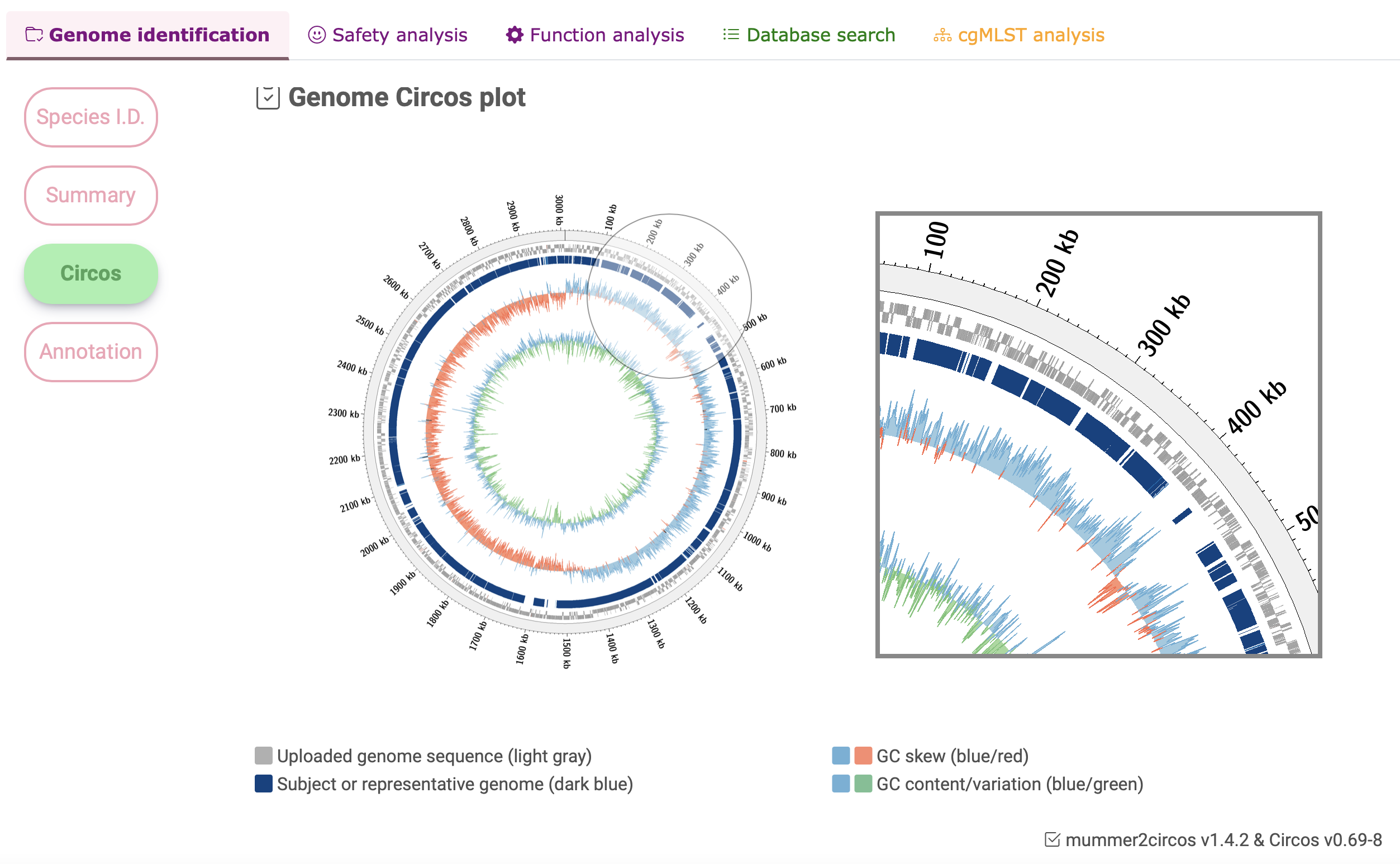
Circos plot
Annotation
Probiotic potential risk score

Antimicrobial resistance gene
Virulence factor
Pathogenic genes

Plasmid
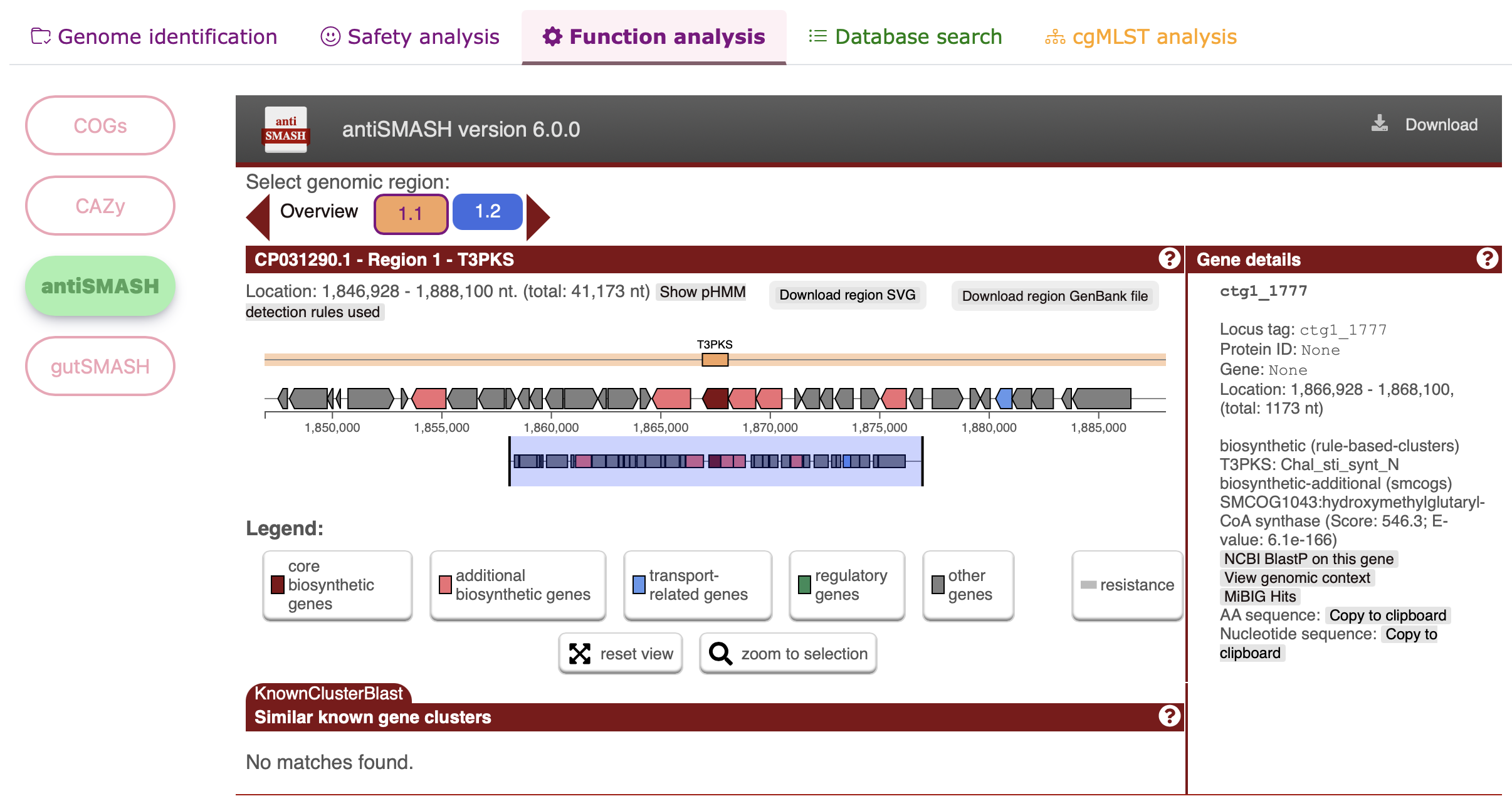
Phigaro
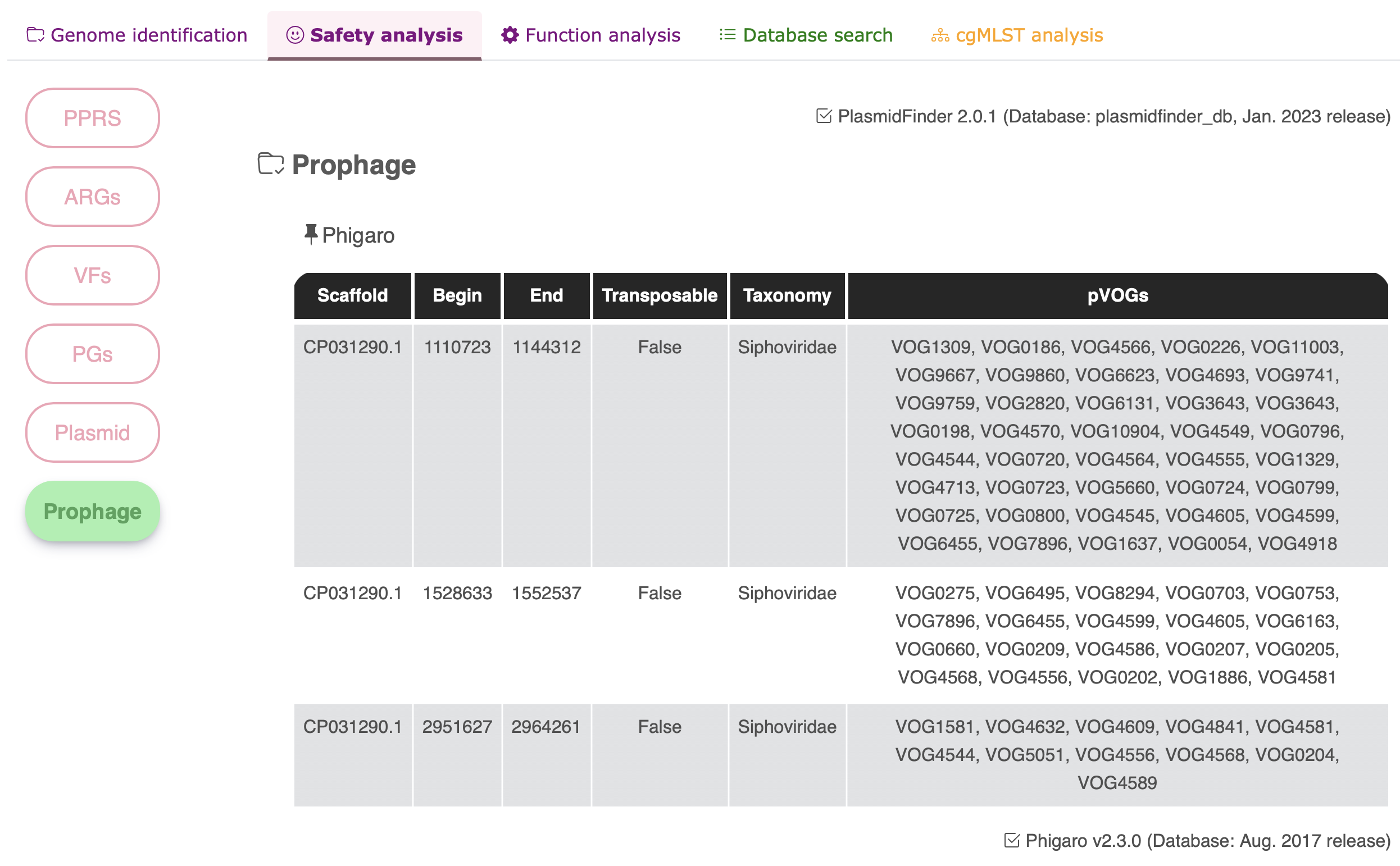
Functional analysis - COGs
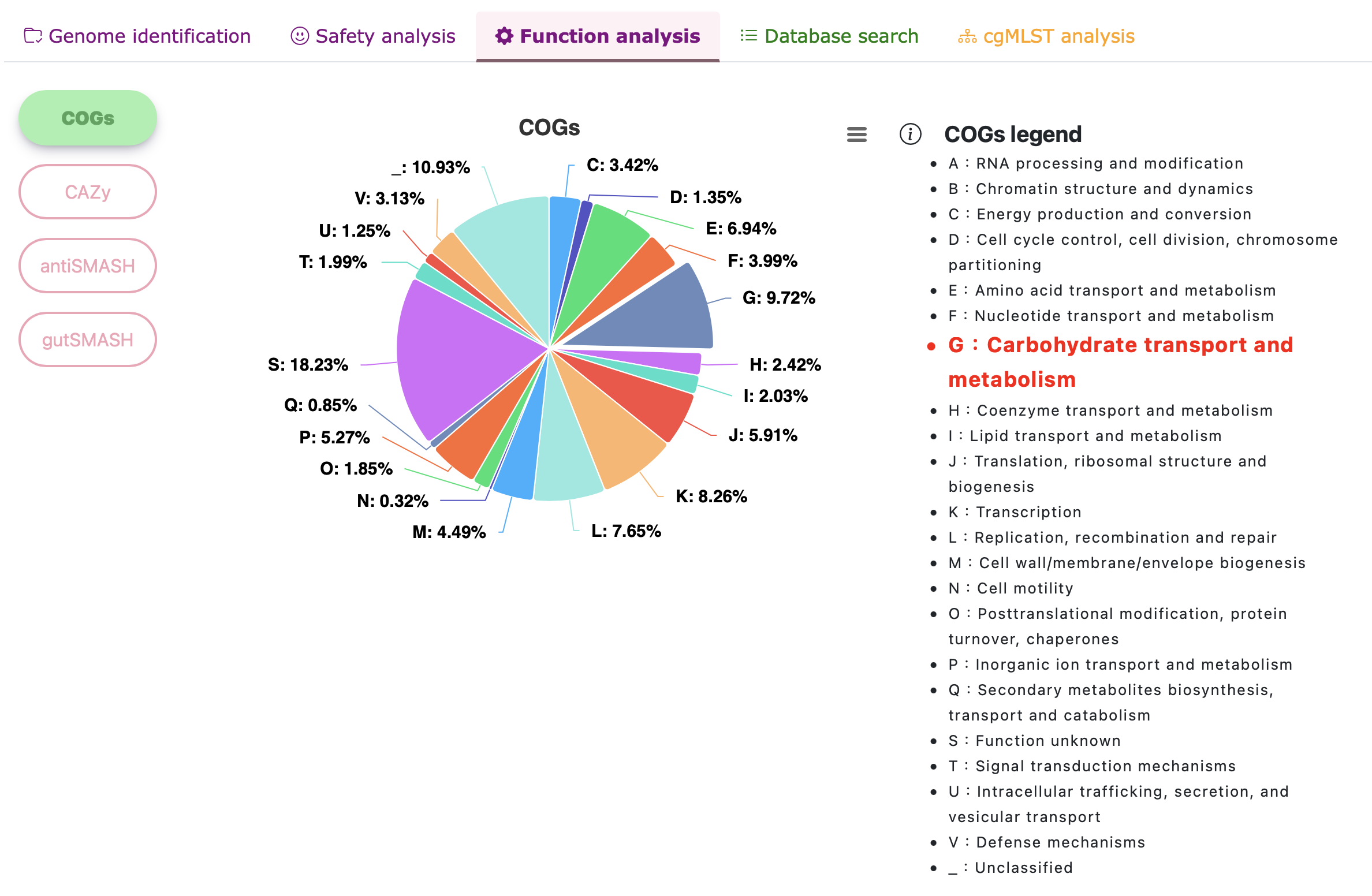
Functional analysis - CAZy
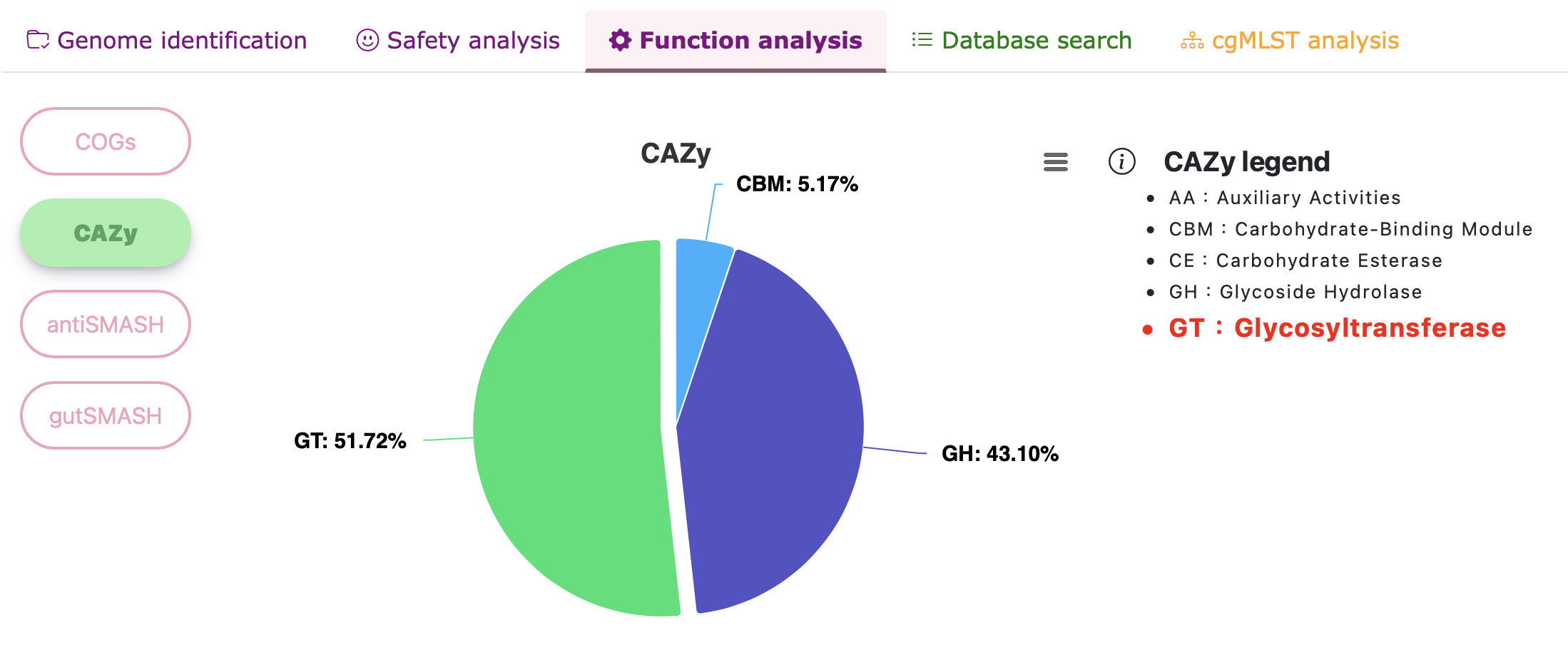
Functional analysis - antiSMASH
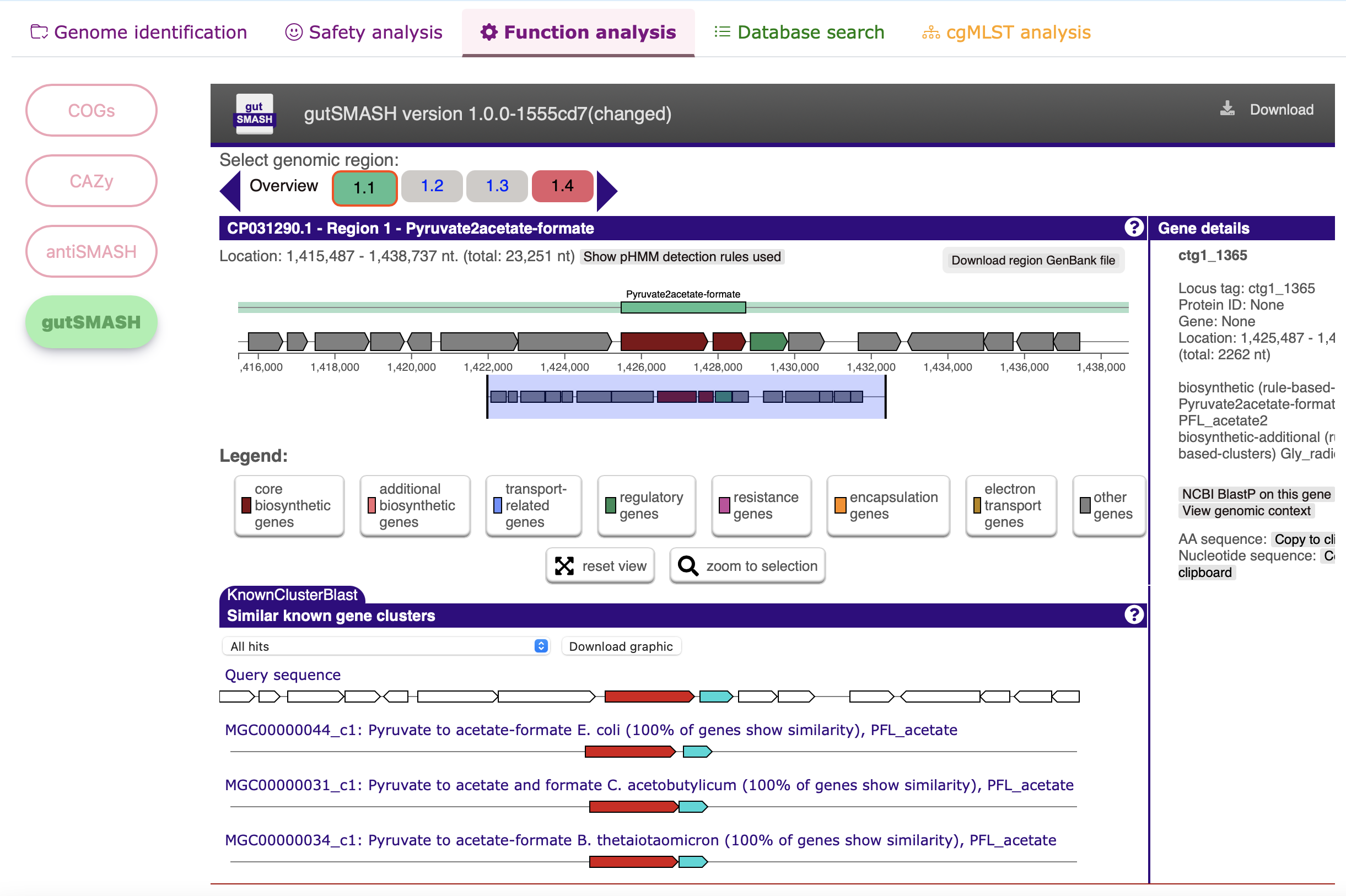
Functional analysis - gutSMASH
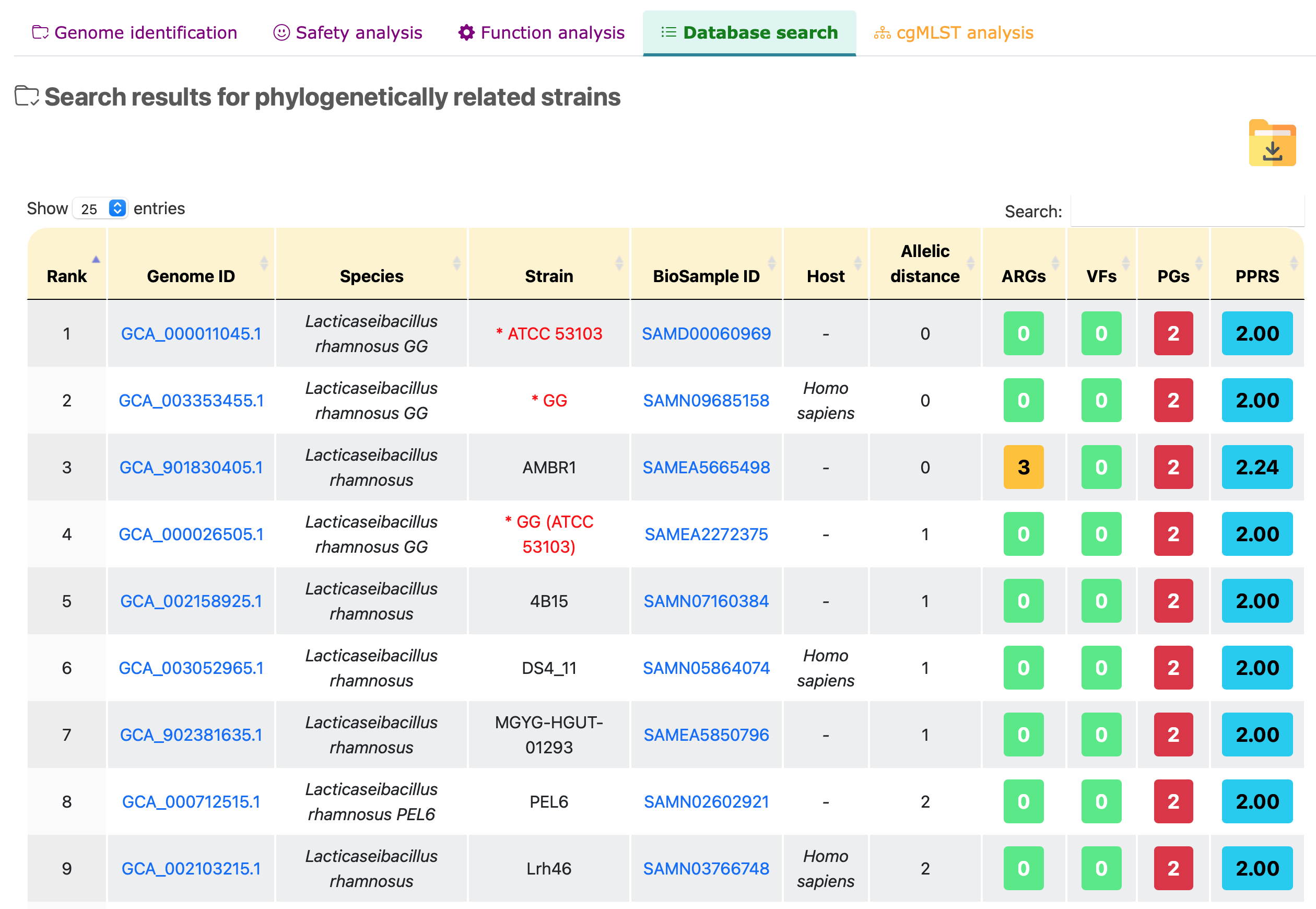
Search results for phylogenetically related strains
cgMLST analysis of top20 hit strains

(IV)List of software and databases used in ProbioMinServer
| Function | Software | Database |
|---|---|---|
| Genome identification | ||
| Type strains comparison | Mash v2.3 | NCBI prokaryote type strains (Aug. 2023 release) |
| Average nucleotide identity (ANI) | https://github.com/chjp/ANI | - |
| Genome Circos plot | mummer2circos v1.4.2 | - |
| Circos v0.69-8 | - | |
| Safety analysis | ||
| Antibiotic resistance genes (ARGs) | RGI v6.0.0 (BLAST homolog detection) | CARD Variants v4.0.0 (July 2022 release) |
| ResFinder v4.0 | resfinder_db (May 2022 release) | |
| AMRFinderPlus v3.10.42 | (October 2022 release) | |
| Virulence factors (VFs) | BLASTN v2.8.1+ | VFDB (June 2022 release) |
| VirulenceFinder v2.0.3 | virulencefinder_db (December 2022 release) | |
| Pathogenic genes (PGs) | BLASTP v2.8.1+ | PHI-base v4.14 (November 2022 release) |
| Plasmid genes | PlasmidFinder v2.0.1 | plasmidfinder_db (January 2023 release) |
| Prophage regions | Phigaro v2.3.0 | (August 2017 release) |
| Functional analysis | ||
| Secondary metabolite gene clusters | antiSMASH v6.0.0 | (June 2022 release) |
| Primary metabolite gene clusters | gutSMASH v1.0.0 | (June 2022 release) |
(V)Browser compatibility
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
|---|---|---|---|---|---|
| Linux | CentOS 7 | not tested | 61.0 | n/a | n/a |
| MacOS | Big Sur, High Sierra | 96.0.4664.110 | 61.0 | n/a | 14.1.1, 13.1.2 |
| Windows | 10 | 96.0.4664.110 | 61.0 | 42.17134.1.0 | n/a |